
 i_need_contribute
i_need_contribute
Doing so would help satisfy a pharma product candidate’s safety criteria: Phase 1 trials. But Phase 2 and Phase 3 (whose purpose is to demonstrate efficacy for the targeted condition) are much harder to assess epidemiologically because COVID-19 has only been with us since December 2019; we just don’t have enough longitudinal data to work with to compare what people are eating and whether or not they recover expediently from infection . . .
In order to jump to the head of the clinical trial line, we want to find 1) people who’ve been exposed to coronaviruses, and 2) find out what plants they might be consuming to manage it. To figure this out, we have one long and circuitous pathway ahead, so buckle up.
For starters, let’s keep in mind how similar that SARS-CoV-2 and SARS-CoV (the one from SARS 2003) viruses are. They’re both coronaviruses (their genomes are 79% identical) — and they both evolved from lineages circulating in bats.
And yet coronaviruses aren’t new to the world — bats have been the reservoir not just for ten or twenty years, not even ten or twenty thousand years, but millions of years. If only we knew which humans were repeatedly exposed to coronaviruses throughout history for our would-be longitudinal study.
To find where such a valuable group of humans can be found, we need to understand a little bit of virology, how virologists go about their work, and crucially, where their work takes them.

(image credit: Iakimchuk Iaroslav | Shutterstock.com)
Virologists study the animal reservoirs of viruses that infect humans. Many viruses are zoonotic: they circulate in animal populations such as bats or birds, before making their way to humans. Often but necessarily always, the virus will evolve its way through an intermediary mammal along the way to human infectability. To study an animal reservoir, the virologists will go wherever any animals suspected of containing the virus live, like the caves of bats.
In caves suspected of harboring coronavirus, bat researchers capture groups of bats at a time. They perform fecal swabs to determine if a bat is infected. They do a PCR test on the samples, similar to tests we are subjected to for COVID-19 today. They then record the virus’ genome as it exists in the bats under study. Stay with me now, because this is where it gets interesting:
Virologists aren’t just looking for any single coronavirus. They are hunting for lineages of the family of coronaviruses. They want to see how the virus varies — how it mutates — from one bat cave to another. Why is that important? They want to see how close any of those coronaviruses are to being capable of hopping to humans.
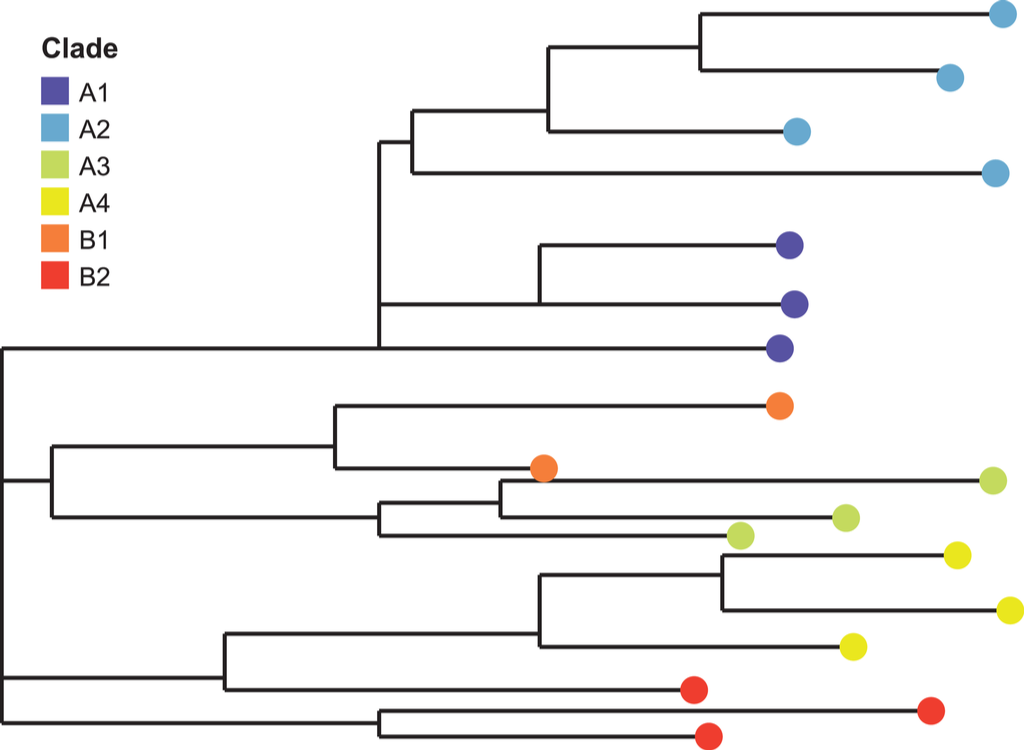
A phylogenetic tree (image credit: M. PATTHAWEE | Shutterstock.com)
To aid virologists in their virus-hunting field work, they employ a tool called a phylogenetic tree. A phylogenetic tree is like a family tree where they compile a listing of each unique version of a virus any researcher has ever encountered and whose genome’s been published. With a phylogenetic tree, the researchers can determine if they are looking at a branch lineage or a parent lineage of a particular viral clade. Virologists are particularly interested in finding a source of viral genetic diversity — one where many related viruses can be found in one place. Such a virally diverse source can indicate the current or historical presence of a parent lineage. It also increases the likelihood that they’ve found the source lineage, and crucially the geographic location, of any virus clade known to have evolved its way to human infectability.
There might be a hundred different clades of coronavirus in circulation, but only a few that are capable of infecting human cells. In order to avoid the next pandemic, you want to know where the bats are that harbor the virus capable of infecting humans. To determine a particular clade’s human infectability, the researchers culture several million virions in a laboratory setting and see if they infect human cells in a test-tube.
The first phase of post-SARS-2003 field research can be thought of as a kind of surveillance: Remember, way back in 2003, the bat origin of coronaviruses was not well-known. All anyone knew was that a mysterious illness started in rural Guangdong and got many people there and in Hong Kong sick. Contact-tracing led them to the earliest cases centered among wildlife traffickers in the Pearl River Delta. The next logical step was to start studying the animals these traffickers traded. And so the coronavirus surveilling began.

(image credit: Jason Mintzer | Shutterstock.com)
Coronavirus researchers swabbed and sampled animals in southern China’s Guangdong and Guangxi provinces. As expected, they found live coronavirus infections and antibody traces of past coronavirus infections among many animals, such as civet cats and other mammals. And bats are just another mammal. So when the researchers started swabbing bats and genotyping the results, they realized that the parent lineages of coronaviruses were in bats. Their phylogenetic tracing led them to the source: neighboring Yunnan province.
But then a curiosity was noted by the bat researchers: Humans living near bat coronavirus hotspots were themselves seropositive for coronavirus antibodies. This evidence of past infections was odd because SARS 2003 was the only coronavirus outbreak known at the time. As it turns out, if you were living in a hotspot area of rural southwest China and had contact with wild animals, it wasn’t an outside possibility that you were once infected with coronavirus at some point in your life.

Image courtesy EcoHealth Alliance (New York)
(At this time it’s probably worth appreciating the amount of work that researchers worldwide, and especially in China, performed in the wake of SARS 2003. A lot of very diligent people labored very hard under very hard-to-get funding. They undertook extensive, painstaking data collection, and published enormous amounts of literature in order to help the world try to manage its way through the next coronavirus pandemic).
The bat coronavirus researchers continued assaying their way up the phylogenetic tree from humans to animals to bats right up to 2015. That was the year they found the cave that was likely the origin of the SARS 2003 virus southwest of the provincial capital. Quite an important find when you think about it. Here is a cave from which so many coronaviruses spring. Between all manner of bat-quadruped-human interactions, these viruses continuously expose, mutate, recombine, and evolve to a point where they’re capable of infecting humans. Finally, these virologists found that local people living near this hallowed cave tested positive for SARSr-CoV (bat) antibodies. However, none could recall SARS-like symptoms, which would make sense if their infection was of a bat-adapted coronavirus.
The research avenue leading to their discovery of the SARS cave made these virologists famous. But their magnum opus announcing their discovery contained one very understated yet very consequential line:
“However, it is noted that two SARSr-CoVs previously reported from Rhinolophus ferrumequinum [greater horseshoe bats] showed the closest phylogenetic position to SARS-CoV (…).
Here’s what that looks like — the clades in question are YNLF_34C and 31C:
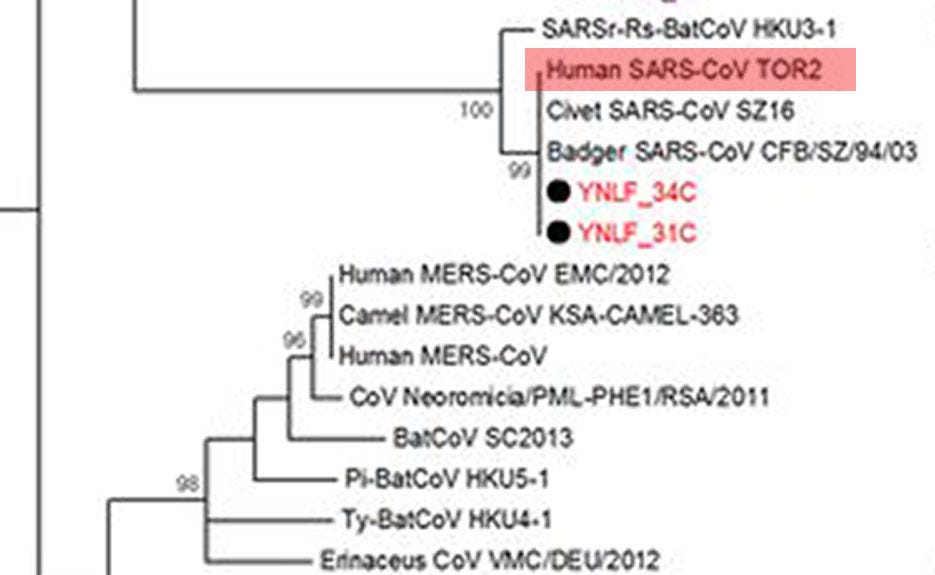
Excerpt of coronavirus phylogenetic tree (Source: Hu et al. “Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus”. Reproduced under fair use.)
and then:
These strains were discovered in another location in Yunnan 80 km from the cave surveyed in the current study.”
In other words, the bat coronavirus clades they recognize as phylogenetically closest to human-infectable SARS are located in a 2nd cave, just 80km to the west. Navigating over to that the paper they are talking about, we are rewarded with more detail:
Phylogenetic analysis (…) of the RdRp gene of betacoronaviruses detected in two bat samples, YNLF_31C and YNLF_34C, showed (…) 100% nucleotide identities to human SARS-CoV TOR2 (. . .) Both samples were collected from greater horseshoe bats (Rhinolophus ferrumequinum) captured in Lufeng County, Chuxiong Yi Autonomous Prefecture.
The cave with the SARSr-CoV clades closest to the human-infectable SARS-CoV is in Lufeng district of Chuxiong Yi prefecture.
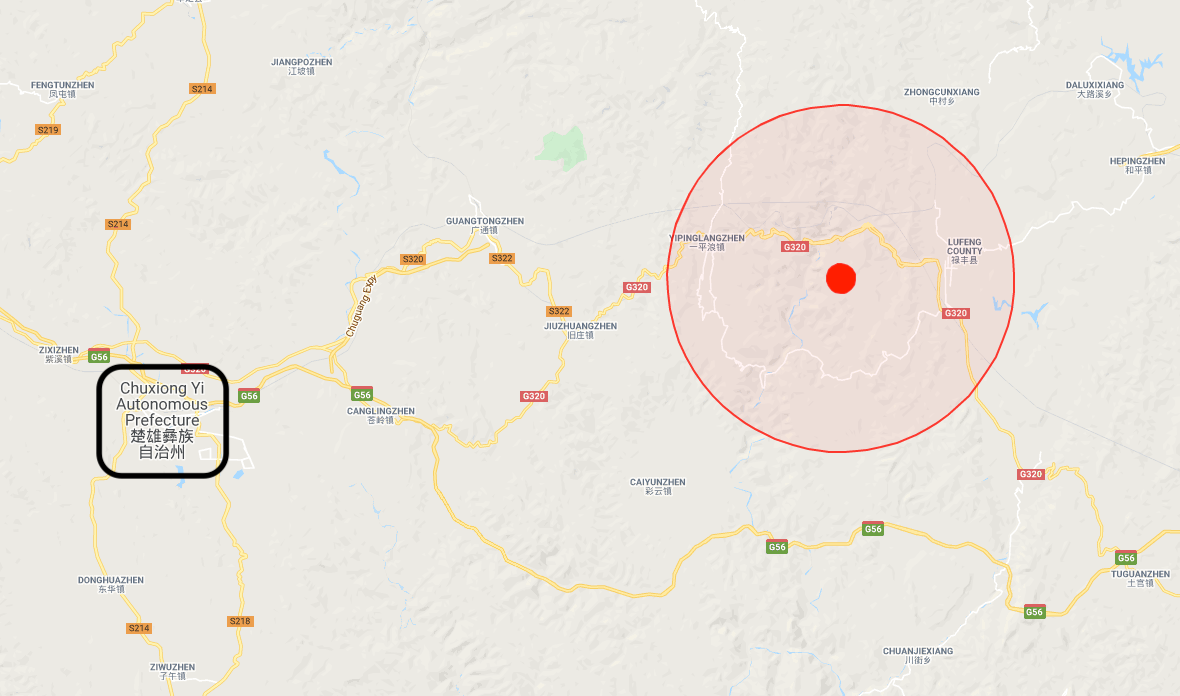
The Lufeng district’s near-human-infectable SARS cave ‘hot zone‘ (Source:EMSKE Phytochem)
Like many other political subdivisions in China, the name of the area indicates the ethnic people predominating in that area — the Yi. In fact, Chuxiong Yi prefecture forms the geographic epicenter for the Yi people.
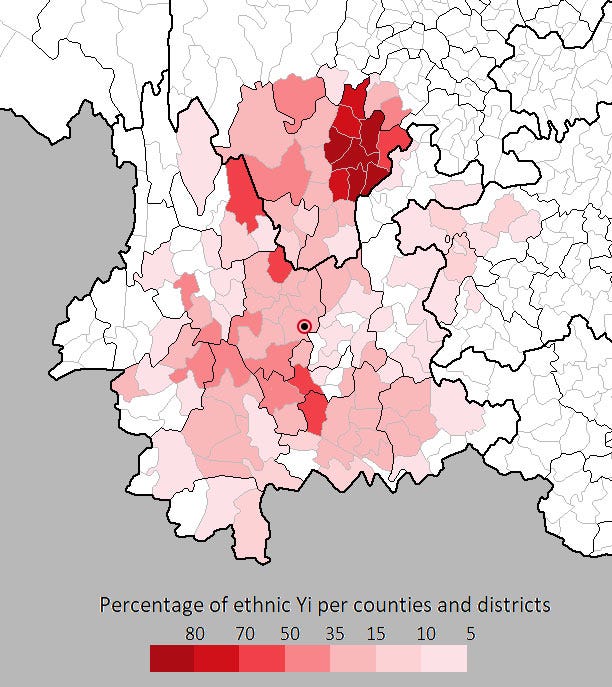
Yi ethnic group in Yunnan — Lufeng cave in Chuxiong Yi prefecture highlighted (by Sgnpkd — Own work with minor EMSKE modification, CC BY-SA 4.0)
It would be reasonable to guess that individuals from this ethnic group living within kilometers of this near-human-infectable SARSr-CoV cave would, if serologically surveilled, manifest antibodies for one clade of coronavirus or another.
We can even surmise that the prefecture’s namesake Yi people who’ve called this area home for millennia have sustained exposure to bat-derived coronaviruses since time immemorial.
But how do we use that information for purposes of generating a longitudinal study? We don’t have Phase I-II-III clinical trial data from these people spanning their centuries of exposure.
Lucky for us, the Yi are multi-generational masters of plant-based traditional medicine . . .

Yi woman in traditional dress (By Zoharby — Own work, CC BY-SA 3.0)
Thank goodness, the Yi’s medicinal system was rigorously inventoried through a series of researcher interviews with traditional Yi herbalists in the same ChuXiong Yi prefecture the cave is located in. Is it possible these people could have learned to treat coronavirus infection long before it ever threatened the world?
The Yi medicinal inventory is organized by which ailments a given herbal medicine treats. Remember, the Yi don’t just come up with these therapeutic indications out of nothing. Rather, they encountered ailments, generation after generation, for hundreds if not thousands of years. Like a living neural network, they’ve learned through hard trial-and-error which herbs work on which ailment and which don’t.

(image credit: Kevin Oh | Shutterstock.com)
Let’s take a moment to appreciate what that means. For a Yi traditional medicine practitioner, finding that an herb didn’t work could very well have meant the loss of that patient — their friend, neighbor, or family member. Repeat that scenario thousands of times over thousands of years, punctuated by the odd success when they got lucky and got it right, and you have yourself a traditional medicine system. Are there dud-indications in any traditional medicine system? Of course. But considering that rigorous traditional medicine analysis was applied to give the world its first-line treatment against malaria (thanks to famous malaria researcher Tu Youyou application of 4th century AD Traditional Chinese Medicine literature), we would be foolish to ignore these systems.

classical Yi written record (credit: smartneddy CC by 3.0)
Taking a page out of Tu Youyou’s book, we scoured the Yi medicinal inventory for symptoms which match the coronavirus symptoms that we all know now all-too-well. As coronavirus is first and foremost a respiratory viral infection, we filter through cough and lung ailment medicinal indications. Three plants are indicated for lung issues, and twelve plants for cough issues. What compounds are in these plants?
Following a lot of subsequent phytochemicals literature sourcing, we end up with a list of over 160 compounds known to be in these 15 plants. We ran these candidate compounds through the same in silico screening tool for SARS-CoV-2 protease inhibition as we used in our recently published assessment of the Artemisia plant extract (‘COVID-Organics’) put out by Madagasar’s applied research institute.
Here are the top-scoring results for coughing indications:
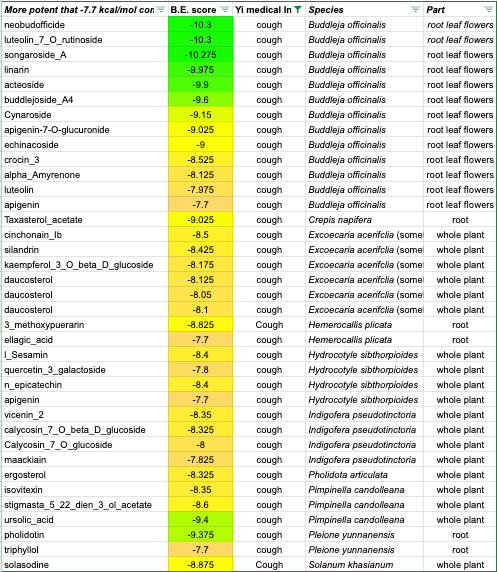
Yi cough indications — constituent compounds top scores (Copyright: EMSKE Phytochem)
And here are the top-scoring results for lung indications:
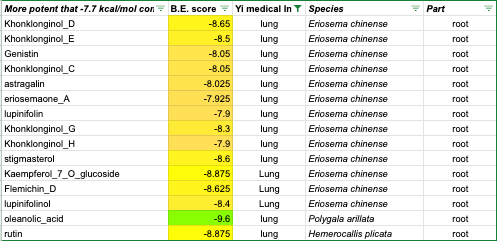
Yi lung indications — constituent compounds top scores (Copyright: EMSKE Phytochem)
As you can see, buddleja officinalis performs astoundingly well. It contains many very high performing hits — right up to the -10 level in our -4 to -12 scale:
acteoside (aka verbascoside): -9.90 kcal/mol
linarin: -9.98 kcal/mol
songoroside A: -10.28 kcal/mol
luteolin-7-O-rutinoside: -10.30 kcal/mol
neobuddoficide: -10.30 kcal/mol
Let’s see what acteoside looks like up against the protease in its highest-binding pose in PyMol. Remember, if the virus is like a bandit in a factory forcing it to make new viruses, then the protease is the virus’ ‘bandsaw’; it cuts freshly-minted peptide chains to size to create new viruses:
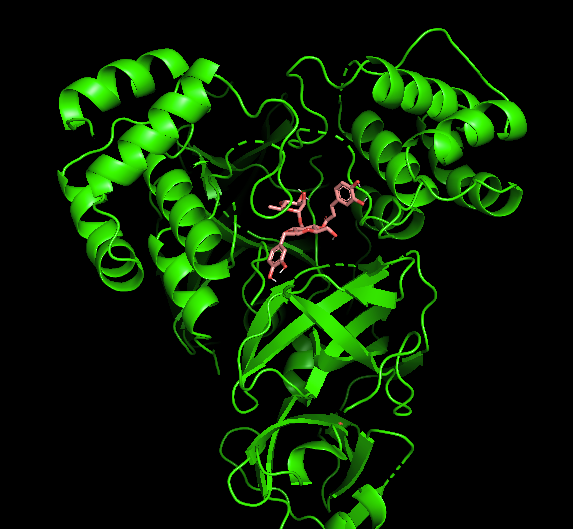

Acteoside binding inside the Mpro’s tunnel structure (left: ‘cartoon’, right:surface) (Image Source: EMSKE Phytochem, Protein: Chen WY, PhD, Program: PyMol)
The compound seats nicely into the protease’s tunnel structure, making binding contact with both symmetrical halves of the protease’s protein machinery. In other words, if you want to throw a wrench into the bandit’s bandsaw, it’s best if it can bind up as much of that machine’s parts as it can.
Three more buddleja compounds, buddlejoside A4, apigenin-7-O-glucuronide, cynaroside, and echinacoside are also considered potent on our scale. And there are still additional, less-potent hits beyond these.
Of course, you can see also that a couple of the other plants seem to perform well. The eriosema chinensis plant matches buddleja for the same number of hits, even if they are less potent ones. The fact that it is specifically indicated for lung issues might render it worthy of further study in its own right. But for scarce clinical trial resources, is there a way of eliminating other plant candidates?
We have to examine the prevalence of high-performing compounds in these plants. Phytochemical assay results for any plant in general span orders of magnitude of weight-for-dry-weight prevalence from compound to compound. And prevalence is a critical stopgap for demonstrating efficacy of a plant extract.

(image courtesy Laurent Renault)
Indeed, the prevalences of most of the high performing compounds listed above are at only trace levels. Not so with buddleja officinalis however. The best information available is that acteoside and linarin are the most prevalent compounds at 6% and 2% of dry weight respectively, with echinacoside coming in at about one-third of linarin prevalence. These values are quite strong by phytochemicals standards. Acteoside’s and linarin’s relative prevalence adds support to these particular compounds’ demonstrated therapeutic value to the Yi. Besides narrowing the field, we actually are getting lucky with these eliminations: There are other species in the buddleja genus that are more widely available than b.officinalis that also contain acteoside and linarin, even while they lack the other high-performing but only trace-level compounds.
The Yi inventory as transcribed unfortunately lacks dosage indications for each plant to a patient. And we don’t have Yi herbalists on speed-dial to learn those dosages.
However, we know from our tool-qualification work that 1) the highest performing compounds require the least concentration to be potent, and 2) that the relationship is logarithmic. So we have a plant possessing 1) several very high-potency compounds, and 2) we have unit-percentage level prevalences of same in the plant’s buds. Therefore we can be assured that the quantities of dry-weight plant material required for efficacy won’t be unreasonable — a gram or two of unrefined plant material extracted in alcohol (per their prescribed preparation method), per dose.
So in short: We assert that the Yi people have likely been treating coronavirus amongst themselves for centuries, and that they’ve been applying buddleja officinalis to do so.
To be useful as a population-scale medicinal over short timescales, there simply has to be enough of the plant to go around. While buddleja is commercially cultivated, it‘s just produced for a low-volume Traditional Chinese Medicine market (for eye-related ailments of all things). So the likelihood of cultivating enough buddleja to be useful to many before the coming northern hemisphere winter is low. Fortunately, the relevant compounds can be found in other even more common plants as well. Where else in commercial crop production might acteoside and linarin be prevalent? We’ll have to save that one for another article . . .
So there we have it: compounds that are known to be
1) safe for consumption at quantities (known to the Yi at least) to be medicinally useful
2) having a very strong prospect of efficacy against coronaviruses in general, with our tool indicating their strong effectiveness in SARS-CoV-2 (COVID-19) inhibition specifically — certainly enough to justify a clinical trial in an animal model, and before very long we hope, trials in a clinical setting.
3) How can we use the lessons of the Yi to be useful at the required scale? Stay tuned for our next article by following us here on Medium or on Twitter at @EMSKEPhyto .
Regarding those bat researchers (and their funders) who labored extensively to help the world manage through the next pandemic: It should be evident that none of the above findings would have been possible without their work from 2004–2018.
The author would like to thank Wai Yu Chen, PhD Protein Crystallography of HK Polytechnic University for his provision of the Mpro protein structure file and instruction in autodocking, and Abdur Azeez, B.S. Biotechnology U. Nottingham for his support in ligand sourcing